Sundance::CellJacobianBatch Class Reference
Public Member Functions | |
| CellJacobianBatch () | |
| int | spatialDim () const |
| int | cellDim () const |
| int | numCells () const |
| int | numQuadPoints () const |
| void | resize (int numCells, int numQuad, int spatialDim, int cellDim) |
| void | resize (int numCells, int spatialDim, int cellDim) |
| double * | jVals (int c, int q) |
| double * | jVals (int c) |
| const double * | jVals (int c) const |
| double * | detJ (int c) |
| const Array< double > & | detJ () const |
| void | applyInvJ (int cell, int q, double *rhs, int nRhs, bool trans) const |
| void | applyInvJ (int cell, double *rhs, int nRhs, bool trans) const |
| void | getInvJ (int cell, int quad, Array< double > &invJ) const |
| void | getInvJ (int cell, Array< double > &invJ) const |
| void | print (std::ostream &os) const |
Static Public Member Functions | |
| static double & | totalFlops () |
| static void | addFlops (const double &flops) |
Private Member Functions | |
| void | factor () const |
| void | computeInverses () const |
Private Attributes | |
| int | spatialDim_ |
| int | cellDim_ |
| int | jSize_ |
| int | numCells_ |
| int | numQuad_ |
| Array< int > | iPiv_ |
| Array< double > | J_ |
| Array< double > | detJ_ |
| Array< double > | invJ_ |
| bool | isFactored_ |
| bool | hasInverses_ |
| bool | hasDetJ_ |
Detailed Description
A CellJacobianBatch is a collection of Jacobian matrices for many quadrature points distributed over batch of cells. All cells must have the same dimension and number of quadrature points. Affine cells have constant Jacobians, so in that case the quadrature point index can be ignored. See the ReferenceCellBase documentation for definitions of the coordinate systems used.
Data layout
All Jacobian elements for all points and cells are packed into a single vector. The length of the vector is 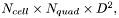 where
where 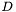 is the spatial dimension. The indices into the vector cycle in the following order (from slowest to fastest)
is the spatial dimension. The indices into the vector cycle in the following order (from slowest to fastest)
- cell number
- quadrature point number
- physical coordinate direction
- reference coordinate direction
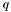 -th quadrature point on the
-th quadrature point on the 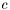 -th cell are the
-th cell are the 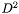 entries following the
entries following the 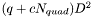 -th element.
-th element.
Definition at line 76 of file SundanceCellJacobianBatch.hpp.
Constructor & Destructor Documentation
| CellJacobianBatch::CellJacobianBatch | ( | ) |
empty ctor
Definition at line 72 of file SundanceCellJacobianBatch.cpp.
Member Function Documentation
| static void Sundance::CellJacobianBatch::addFlops | ( | const double & | flops | ) | [inline, static] |
Definition at line 162 of file SundanceCellJacobianBatch.hpp.
References totalFlops().
Referenced by applyInvJ(), computeInverses(), factor(), Sundance::BasicSimplicialMesh::getJacobians(), and Sundance::BasicSimplicialMesh::pushForward().
| void Sundance::CellJacobianBatch::applyInvJ | ( | int | cell, | |
| double * | rhs, | |||
| int | nRhs, | |||
| bool | trans | |||
| ) | const [inline] |
Apply an affine cell's inverse Jacobian to (possibly) multiple rhs stored in column-major order.
Definition at line 137 of file SundanceCellJacobianBatch.hpp.
References applyInvJ().
Referenced by applyInvJ().
| void CellJacobianBatch::applyInvJ | ( | int | cell, | |
| int | q, | |||
| double * | rhs, | |||
| int | nRhs, | |||
| bool | trans | |||
| ) | const |
Apply a cell's inverse Jacobian to (possibly) multiple rhs stored in column-major order.
Definition at line 215 of file SundanceCellJacobianBatch.cpp.
References addFlops(), dgetrs_(), factor(), iPiv_, isFactored_, J_, numCells_, numQuad_, and spatialDim_.
Referenced by Sundance::CurveIntegralCalc::getCurveQuadPoints_line(), and Sundance::CurveIntegralCalc::getCurveQuadPoints_polygon().
| int Sundance::CellJacobianBatch::cellDim | ( | ) | const [inline] |
get the cell dimension
Definition at line 87 of file SundanceCellJacobianBatch.hpp.
References cellDim_.
Referenced by Sundance::ElementIntegral::createOneFormTransformationMatrix(), Sundance::ElementIntegral::createTwoFormTransformationMatrix(), detJ(), and Sundance::CurveEvalMediator::evalDiscreteFuncElement().
| void CellJacobianBatch::computeInverses | ( | ) | const [private] |
Definition at line 168 of file SundanceCellJacobianBatch.cpp.
References addFlops(), dgetrs_(), factor(), hasInverses_, invJ_, iPiv_, isFactored_, J_, jacobianInversionTimer(), jSize_, numCells_, numQuad_, spatialDim_, SUNDANCE_OUT, and Playa::ObjectWithVerbosity::verb().
| const Array<double>& Sundance::CellJacobianBatch::detJ | ( | ) | const [inline] |
get the vector of determinant values
Definition at line 123 of file SundanceCellJacobianBatch.hpp.
References cellDim(), detJ_, factor(), isFactored_, and spatialDim().
Referenced by factor().
| double* Sundance::CellJacobianBatch::detJ | ( | int | c | ) | [inline] |
Definition at line 119 of file SundanceCellJacobianBatch.hpp.
References detJ_.
Referenced by Sundance::ElementIntegral::createOneFormTransformationMatrix(), Sundance::ElementIntegral::createTwoFormTransformationMatrix(), Sundance::PeriodicSingleCellMesh1D::getJacobians(), Sundance::PeriodicMesh1D::getJacobians(), Sundance::HNMesh3D::getJacobians(), Sundance::HNMesh2D::getJacobians(), Sundance::BasicSimplicialMesh::getJacobians(), Sundance::RefIntegral::transformOneForm(), Sundance::ReducedIntegral::transformOneForm(), Sundance::QuadratureIntegral::transformOneForm(), Sundance::MaximalQuadratureIntegral::transformOneForm(), Sundance::CurveQuadratureIntegral::transformOneForm(), Sundance::CurveQuadratureIntegral::transformSummingFirst(), Sundance::RefIntegral::transformTwoForm(), Sundance::ReducedIntegral::transformTwoForm(), Sundance::QuadratureIntegral::transformTwoForm(), Sundance::MaximalQuadratureIntegral::transformTwoForm(), Sundance::RefIntegral::transformZeroForm(), Sundance::ReducedIntegral::transformZeroForm(), Sundance::QuadratureIntegral::transformZeroForm(), and Sundance::MaximalQuadratureIntegral::transformZeroForm().
| void CellJacobianBatch::factor | ( | ) | const [private] |
Definition at line 115 of file SundanceCellJacobianBatch.cpp.
References addFlops(), cellDim_, detJ(), detJ_, dgetrf_(), iPiv_, isFactored_, J_, jacobianFactoringTimer(), jSize_, numCells_, numQuad_, spatialDim_, SUNDANCE_OUT, and Playa::ObjectWithVerbosity::verb().
Referenced by applyInvJ(), computeInverses(), and detJ().
| void Sundance::CellJacobianBatch::getInvJ | ( | int | cell, | |
| Array< double > & | invJ | |||
| ) | const [inline] |
Get the explicit inverse of the Jacobian for the given affine cell.
Definition at line 151 of file SundanceCellJacobianBatch.hpp.
References getInvJ().
Referenced by getInvJ().
| void CellJacobianBatch::getInvJ | ( | int | cell, | |
| int | quad, | |||
| Array< double > & | invJ | |||
| ) | const |
Get the explicit inverse of the Jacobian for the given (cell, quad) combination.
Definition at line 241 of file SundanceCellJacobianBatch.cpp.
References computeInverses(), hasInverses_, invJ_, jSize_, numQuad_, and spatialDim_.
Referenced by Sundance::ElementIntegral::createOneFormTransformationMatrix(), Sundance::ElementIntegral::createTwoFormTransformationMatrix(), and Sundance::CurveEvalMediator::evalDiscreteFuncElement().
| const double* Sundance::CellJacobianBatch::jVals | ( | int | c | ) | const [inline] |
Get a constant pointer to start of c-th Jacobian in the batch
Definition at line 116 of file SundanceCellJacobianBatch.hpp.
| double* Sundance::CellJacobianBatch::jVals | ( | int | c | ) | [inline] |
Get a pointer to the start of the c-th Jacobian in the batch.
Definition at line 112 of file SundanceCellJacobianBatch.hpp.
| double* Sundance::CellJacobianBatch::jVals | ( | int | c, | |
| int | q | |||
| ) |
Get a pointer to the values at the q-th quadrature point on the c-th cell.
- Parameters:
-
c the index of the cell in the batch q the index of the quadrature point
Referenced by Sundance::PeriodicSingleCellMesh1D::getJacobians(), Sundance::PeriodicMesh1D::getJacobians(), Sundance::HNMesh3D::getJacobians(), Sundance::HNMesh2D::getJacobians(), Sundance::BasicSimplicialMesh::getJacobians(), Sundance::CubicHermite::postApplyTransformationTriangle(), Sundance::CubicHermite::preApplyTransformationTransposeTriangle(), and Sundance::CubicHermite::preApplyTransformationTriangle().
| int Sundance::CellJacobianBatch::numCells | ( | ) | const [inline] |
get the number of cells in the batch
Definition at line 90 of file SundanceCellJacobianBatch.hpp.
References numCells_.
Referenced by Sundance::ElementIntegral::createOneFormTransformationMatrix(), Sundance::ElementIntegral::createTwoFormTransformationMatrix(), Sundance::CurveEvalMediator::evalDiscreteFuncElement(), Sundance::IntegralGroup::evaluate(), Sundance::CubicHermite::postApplyTransformationTriangle(), Sundance::CubicHermite::preApplyTransformationTransposeTriangle(), Sundance::CubicHermite::preApplyTransformationTriangle(), Sundance::RefIntegral::transformOneForm(), Sundance::ReducedIntegral::transformOneForm(), Sundance::QuadratureIntegral::transformOneForm(), Sundance::MaximalQuadratureIntegral::transformOneForm(), Sundance::CurveQuadratureIntegral::transformOneForm(), Sundance::CurveQuadratureIntegral::transformSummingFirst(), Sundance::RefIntegral::transformTwoForm(), Sundance::ReducedIntegral::transformTwoForm(), Sundance::QuadratureIntegral::transformTwoForm(), Sundance::MaximalQuadratureIntegral::transformTwoForm(), Sundance::CurveQuadratureIntegral::transformTwoForm(), Sundance::RefIntegral::transformZeroForm(), Sundance::ReducedIntegral::transformZeroForm(), Sundance::QuadratureIntegral::transformZeroForm(), Sundance::MaximalQuadratureIntegral::transformZeroForm(), and Sundance::CurveQuadratureIntegral::transformZeroForm().
| int Sundance::CellJacobianBatch::numQuadPoints | ( | ) | const [inline] |
get the number of quad points per cell
Definition at line 93 of file SundanceCellJacobianBatch.hpp.
References numQuad_.
| void CellJacobianBatch::print | ( | std::ostream & | os | ) | const |
Definition at line 258 of file SundanceCellJacobianBatch.cpp.
References computeInverses(), hasInverses_, invJ_, jSize_, numCells_, numQuad_, and spatialDim_.
Referenced by Sundance::operator<<().
| void CellJacobianBatch::resize | ( | int | numCells, | |
| int | spatialDim, | |||
| int | cellDim | |||
| ) |
resize the batch, using one quadrature point per cell (appropriate for affine elements)
Definition at line 97 of file SundanceCellJacobianBatch.cpp.
References cellDim_, detJ_, hasInverses_, iPiv_, isFactored_, J_, jSize_, numCells_, numQuad_, and spatialDim_.
| void CellJacobianBatch::resize | ( | int | numCells, | |
| int | numQuad, | |||
| int | spatialDim, | |||
| int | cellDim | |||
| ) |
resize the batch
Definition at line 78 of file SundanceCellJacobianBatch.cpp.
References cellDim_, detJ_, hasInverses_, iPiv_, isFactored_, J_, jSize_, numCells_, numQuad_, and spatialDim_.
Referenced by Sundance::CurveIntegralCalc::getCurveQuadPoints_line(), Sundance::CurveIntegralCalc::getCurveQuadPoints_polygon(), Sundance::PeriodicSingleCellMesh1D::getJacobians(), Sundance::PeriodicMesh1D::getJacobians(), Sundance::HNMesh3D::getJacobians(), Sundance::HNMesh2D::getJacobians(), and Sundance::BasicSimplicialMesh::getJacobians().
| int Sundance::CellJacobianBatch::spatialDim | ( | ) | const [inline] |
get the spatial dimension
Definition at line 84 of file SundanceCellJacobianBatch.hpp.
References spatialDim_.
Referenced by detJ(), and Sundance::CurveEvalMediator::evalDiscreteFuncElement().
| static double& Sundance::CellJacobianBatch::totalFlops | ( | ) | [inline, static] |
Member Data Documentation
int Sundance::CellJacobianBatch::cellDim_ [private] |
Definition at line 171 of file SundanceCellJacobianBatch.hpp.
Array<double> Sundance::CellJacobianBatch::detJ_ [mutable, private] |
Definition at line 177 of file SundanceCellJacobianBatch.hpp.
bool Sundance::CellJacobianBatch::hasDetJ_ [mutable, private] |
Definition at line 181 of file SundanceCellJacobianBatch.hpp.
bool Sundance::CellJacobianBatch::hasInverses_ [mutable, private] |
Definition at line 180 of file SundanceCellJacobianBatch.hpp.
Referenced by computeInverses(), getInvJ(), print(), and resize().
Array<double> Sundance::CellJacobianBatch::invJ_ [mutable, private] |
Definition at line 178 of file SundanceCellJacobianBatch.hpp.
Referenced by computeInverses(), getInvJ(), and print().
Array<int> Sundance::CellJacobianBatch::iPiv_ [mutable, private] |
Definition at line 175 of file SundanceCellJacobianBatch.hpp.
Referenced by applyInvJ(), computeInverses(), factor(), and resize().
bool Sundance::CellJacobianBatch::isFactored_ [mutable, private] |
Definition at line 179 of file SundanceCellJacobianBatch.hpp.
Referenced by applyInvJ(), computeInverses(), detJ(), factor(), and resize().
Array<double> Sundance::CellJacobianBatch::J_ [mutable, private] |
Definition at line 176 of file SundanceCellJacobianBatch.hpp.
Referenced by applyInvJ(), computeInverses(), factor(), jVals(), and resize().
int Sundance::CellJacobianBatch::jSize_ [private] |
Definition at line 172 of file SundanceCellJacobianBatch.hpp.
Referenced by computeInverses(), factor(), getInvJ(), jVals(), print(), and resize().
int Sundance::CellJacobianBatch::numCells_ [private] |
Definition at line 173 of file SundanceCellJacobianBatch.hpp.
Referenced by applyInvJ(), computeInverses(), factor(), numCells(), print(), and resize().
int Sundance::CellJacobianBatch::numQuad_ [private] |
Definition at line 174 of file SundanceCellJacobianBatch.hpp.
Referenced by applyInvJ(), computeInverses(), factor(), getInvJ(), numQuadPoints(), print(), and resize().
int Sundance::CellJacobianBatch::spatialDim_ [private] |
Definition at line 170 of file SundanceCellJacobianBatch.hpp.
Referenced by applyInvJ(), computeInverses(), factor(), getInvJ(), print(), resize(), and spatialDim().


